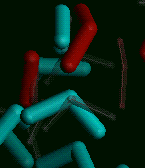 Pre-recorded Script Lesson,
Instructions
Pre-recorded Script Lesson,
Instructions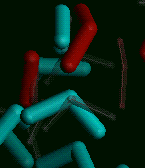 Pre-recorded Script Lesson,
Instructions
Pre-recorded Script Lesson,
InstructionsIssue the command biosym_tutorial -i at the system prompt. If the Discover tutorial files are not installed, enter discover at the first prompt.
Additional information on installing tutorials.
Press <Enter> to get out of the installation menu, then enter 3 to change to the tutorial directory, followed by entering 1 to start up the Insight II program.
It takes a few moments for the Insight program to start up.
Select the File/Import command. Choose the file name diffusion.car from the Parameters value-aid. Execute the command.
A collection of water molecules appears in the display area of the Insight screen. You may select Cancel to put away the parameter block so you can see the molecules better.
Select the Object/LineWidth command. Set the Object Name parameter to DIFFUSION by choosing DIFFUSION from the value-aid (if necessary). In the Line Width parameter box, enter 3. Select Execute or press <Enter>.
Select the Molecule/Color command. The Molecule Spec parameter box should contain COPY; if it does not, choose COPY from the value-aid. Click the Color parameter box and enter 100,100,100. Select Execute.
Select the Object/LineWidth command. Click the Line Width parameter box and enter 1. Select Execute.
The copy molecules are now colored dark gray and are displayed with narrow lines, to make them less noticeable than the original molecules, as the original molecules move during the subsequent Discover run.
Select the Molecule/Color command. Change the Molecule Spec parameter box by choosing DIFFUSION from the value-aid. Pick the cyan square from the palette to fill in the Color parameter box (or enter 0,255,255). Select Execute.
The original molecules are now colored cyan.
The precoded Discover 95.0 simulation we will be running will "heat" atoms within 5 Å of the center of the periodic cell; we will highlight those atoms by coloring them red:
Select the Subset/Delete command. Fill in the Subset Name parameter box by choosing HOT from the value-aid. Select Execute.
Go to the Module pulldown (that is, click the Biosym logo) and select Discover_3 from the list that appears.
The Discover_3 pulldowns appear on the lower menu bar.
Choose DIFFUSION from the value-aid. This enters DIFFUSION in the Disco Object Name parameter box of the command. Select Execute.
This action informs the Discover system that you want to work on the collection of water molecules that is named DIFFUSION.
Select the Strategy/Use_Existing_Input command. Fill in the Object_Name parameter box by choosing DIFFUSION from the value-aid. Click the Input_ File parameter box and choose diffusion.inp from the value-aid. Select Execute.
Once the ``Working'' label changes back to Execute, you may select Cancel to put away the parameter block.
The job begins to run (it is not necessary to use the D_Run/Run command to start the job when running an existing strategy file). After a few seconds of setup time (depending on the speed of your processor and the number of other processes running at the same time), the coordinates of the original water molecules begin to be updated periodically as the interactive simulation proceeds. You can observe that the red atoms are moving much further at each step than the cyan atoms; this is because the precoded Discover 95.0 script has simulated heating the red atoms, so their velocities are much greater than those of the cyan atoms.
The simulation runs for 400 steps (100 fs with a timestep of 0.25 fs) of dynamics. If you want to stop it early, you can use the Background_Job/Kill_Bkgd_Job command.
If you allow the job to run to completion, a notifier window appears. Select the Continue button in the notifier window. You may now compare your output file with an annotated copy of the output file:
To examine the output file, select the Session/Unix command and select Execute. Wait until a shell window and a UNIX prompt appear, then list the output file by entering the following command at the UNIX prompt (represented by >):
> more DIFFUSION1.outAfter viewing the output file, return to the Insight environment by typing exit at the UNIX prompt and then selecting the Textport button.
Type quit on in the command line near the bottom of the Insight screen and press <Enter>.
Now enter 5 to exit the biosym_tutorial script.
 Main
access page
Main
access page  Insight UIF access
Insight UIF access
 Insight UIF - Tutorial access.
Insight UIF - Tutorial access.
 Pre-recorded Script Lesson, Introduction
Pre-recorded Script Lesson, Introduction