 Minimization Lesson,
Instructions
Minimization Lesson,
Instructions Minimization Lesson,
Instructions
Minimization Lesson,
InstructionsAdditional information on installing tutorials.
Press <Enter> to get out of the installation menu, then enter 3 to change to the tutorial directory, followed by entering 1 to start up the Insight II program.
It takes a few moments for the Insight program to start up.
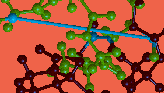
The molecule that appears was constructed by forming a (distorted) bond between the first and last residues of the linear peptide gly-pro-gly-ala-arg.
You can use the mouse buttons to translate and rotate the molecule to facilitate viewing it.
Go to the Module pulldown (that is, click the Biosym logo) and select Discover_3 from the list that appears.
The Discover_3 pulldowns appear on the lower menu bar.
Pick the peptide structure in the display area or choose CYCLO0 from the Assem/Mol Names value-aid to fill in the Disco Object Name parameter box.
Select Execute.
This defines CYCLO0 as the system that you want to work with and also enables you to execute additional Discover_3 commands.
At any time after this step, you may compare your input file with an annotated copy of the input file, by going to a different shell window (i.e., not the one from which you started the Insight program) and using the UNIX cd command to go to your tutorial directory, then the more command to list the cyclo00.inp file.
The conformation of the molecule is updated as the run proceeds, and information on the progress of the run is printed in the display area.
When the job is finished (less than a minute on a 100-MHz SGI Indigo2 IRIX 5.2) a window appears, indicating that the job has completed. Click the Continue button to put the window away.
Select the File/Import command from the upper menu bar. Specify cyclo0.car as the value of the File_Name parameter and Execute the command.
This brings the original structure into the Insight display area (probably on top of the minimized structure).
Connect the mouse to either of the molecules (use the Connect Object button) to move it without moving the other. Move the molecules around to compare their configurations--note that the long bond in the original molecule is now indistinguishable in length from the other bonds in the optimized molecule.
> more cyclo00.outYou may compare your output file with an annotated copy of the output file.
Note that the default calculation uses group-based nonbond energy cutoffs of 9.50 Å for both the van der Waals and Coulombic interactions.
After viewing the output file, return to the Insight environment by typing exit at the UNIX prompt and then selecting the Textport button.
This completes the illustration of a simple minimization.
Select the Object/Delete command from the upper menu bar. Specify the original structure (now called CYCLO01) and select Execute.
 Main
access page
Main
access page  Insight UIF access
Insight UIF access
 Insight UIF - Tutorial access.
Insight UIF - Tutorial access.
 Minimization Lesson, Introduction
Minimization Lesson, Introduction
 Simple Dynamics Lesson, Introduction
Simple Dynamics Lesson, Introduction
Copyright Biosym/MSI