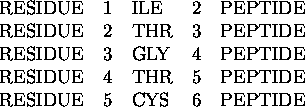
Models containing only a single chain are very simple to describe. The sequence file consists of a list of residues in the model, each with its own type and links. All of this information is provided on RESIDUE statements. They have the following form
RESIDUE <Residue name> <Residue type> -
N(<Residue name> <Link type>)
There will be one RESIDUE statement for each residue in the model. Following the keyword `RESIDUE' is the name of the residue. This is followed by its type. Each residue must have a unique name
The last part of the RESIDUE card specifies the target and link type of any connections between this residue and any other residues.
In the following example residue ``1'', an isoleucine, is connected to residue ``2'' by a peptide bond (named PEPTIDE). Residue ``2'', threonine, is connected to residue ``3'' by a peptide bond. Residue ``3'', glycine, is connected to residue ``4'' by a peptide bond, and so on.
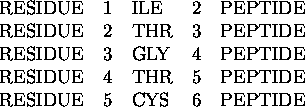
Because residues are recognized by names and not numbers, ``numbers'' may be left out of the sequence, and additional residues may be inserted once the sequence has been numbered, as shown in the next example.

A disulfide bond is shown in the following example. It connects residues 13 and 67.
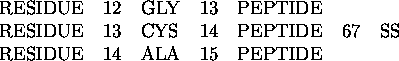
There are two ways to terminate a polypeptide chain. The linkage type CTERM is used at a normal C-terminus. The extra oxygen of the carboxyl acid group is placed in a separate residue and the two final residues are linked with CTERM. Often a peptide model comes to an end not because of chemistry, but because the density becomes so weak that the chain can no longer be traced reliably. In this case there would not necessarily be an extra oxygen atom -- the chain simply ends abruptly.
Because of the way the geometry of amino acids is defined all amino acids must be linked to something on their C-terminal end. If the model stops the last amino acid is linked to something else (anything else) with a BREAK link (I know, ``BREAK link" sounds a little strange but you try to think all this stuff up.). The target of this link could be the amino acid on the far side of the gap or just a dummy residue created simply to be a target.
The final example shows how to include a break, a C-terminus, metal ions, and a heme group in the connectivity file.

In this example, the protein contains four calcium ions, one zinc ion, and a heme. There is a break in the chain between residues ``306'' and ``314''. The residue ``NULL'' has been created simply to have something for ``306'' to be connected. The additional oxygen atom present at the C-terminus is treated as a separate group with the name ``COOH'' which is bonded to the C-terminal residue with the geometry specified for ``CTERM''.