More example images can be found in the galleries of the Official MolScript Web Site.
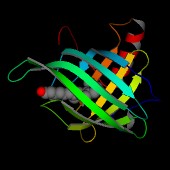 This image shows the secondary structure coloured in a rainbow ramp
from blue at the N-terminus to red at the C-terminus. The retinol
molecule is shown in a CPK representation.
This image shows the secondary structure coloured in a rainbow ramp
from blue at the N-terminus to red at the C-terminus. The retinol
molecule is shown in a CPK representation.
% molscript -jpeg 90 -accum 4 -size 170 170 -out rbp-std.jpg < rbp-std.in
!----------------------------------------------------------------
! MolScript v2.0 input file rbp-std.in
!
! Initial file produced by MolAuto v1.0, then modified by hand.
!
! RBP, standard view.
!
! Per Kraulis
! 22-Oct-1997
title "Retinol-binding protein"
plot
slab 50;
read mol "pdb1rbp.ent";
transform atom * by centre position atom *
by rotation
-0.593897 0.165647 -0.787304
0.371135 0.924642 -0.0854205
0.713824 -0.342927 -0.610619
by translation 4 0 0
by rotation x -6
;
set segments 6;
set planecolour hsb 0.6667 1 1;
coil from 1 to 7;
helix from 7 to 9;
coil from 9 to 22;
set planecolour hsb 0.5333 1 1;
strand from 22 to 30;
coil from 30 to 39;
set planecolour hsb 0.4667 1 1;
strand from 39 to 47;
coil from 47 to 53;
set planecolour hsb 0.4 1 1;
strand from 53 to 62;
coil from 62 to 68;
set planecolour hsb 0.3333 1 1;
strand from 68 to 79;
coil from 79 to 85;
set planecolour hsb 0.2667 1 1;
strand from 85 to 92;
coil from 92 to 100;
set planecolour hsb 0.2 1 1;
strand from 100 to 109;
coil from 109 to 114;
set planecolour hsb 0.1333 1 1;
strand from 114 to 120;
coil from 120 to 132;
set planecolour hsb 0.06667 1 1;
strand from 132 to 138;
coil from 138 to 146;
set planecolour hsb 0 1 1;
helix from 146 to 158;
coil from 158 to 175;
set atomcolour atom C* grey 0.5;
cpk in require residue 1 and type RTL;
end_plot
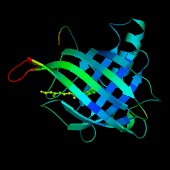 This image shows the secondary structure coloured according to the
B-factor values of the CA atom in each residue. The colours go from
blue (B-factor 8.0), through green to red (B-factor 45.0).
The retinol molecule is shown in a ball-and-stick representation with
the atoms also coloured according to their respective B-factors.
This image shows the secondary structure coloured according to the
B-factor values of the CA atom in each residue. The colours go from
blue (B-factor 8.0), through green to red (B-factor 45.0).
The retinol molecule is shown in a ball-and-stick representation with
the atoms also coloured according to their respective B-factors.
% molscript -jpeg 90 -accum 4 -size 170 170 -out rbp-bfactor.jpg < rbp-bfactor.in
!----------------------------------------------------------------
! MolScript v2.0 input file rbp-bfactor.in
!
! Initial file produced by MolAuto v1.0, then modified by hand.
!
! RBP, B-factor colours.
!
! Per Kraulis
! 22-Oct-1997
title "Retinol-binding protein"
plot
slab 50;
read mol "pdb1rbp.ent";
transform atom * by centre position atom *
by rotation
-0.593897 0.165647 -0.787304
0.371135 0.924642 -0.0854205
0.713824 -0.342927 -0.610619
by translation 4 0 0
by rotation x -6
;
set segments 6;
set hsbrampreverse on,
residuecolour residue * b-factor 8 45 from blue to red,
atomcolour atom * b-factor 8 45 from blue to red,
colourparts on;
coil from 1 to 7;
helix from 7 to 9;
coil from 9 to 22;
strand from 22 to 30;
coil from 30 to 39;
strand from 39 to 47;
coil from 47 to 53;
strand from 53 to 62;
coil from 62 to 68;
strand from 68 to 79;
coil from 79 to 85;
strand from 85 to 92;
coil from 92 to 100;
strand from 100 to 109;
coil from 109 to 114;
strand from 114 to 120;
coil from 120 to 132;
strand from 132 to 138;
coil from 138 to 146;
helix from 146 to 158;
coil from 158 to 175;
ball-and-stick in require residue 1 and type RTL;
end_plot